Pattern
Beside the regular pattern, which result from identical base sequences that repeat after a constant number of bases there also can be seen slight structures in the pictures which are spread all over the whole chromosomes. A change in the width of our images results in images with other but similar structures all over the chromosome. This leads to the suggestion that the structures result from a stochastic distribution of the colors given to each pixel as in a pure random image.
Although the structures are distributed over wide chromosomal areas containing “Junk DNA” with no obvious actual function for the living organism, they also can be noticed in areas containing many genes. Since each color in our images stands for one of the four bases of the DNA, in these gene-rich areas the sequence of the bases/colors is definitely not of stochastic nature. Thus, one might explain the structures as an irregular repeat of similar base sequences. If the structures encode for any information is unclear.
Image enhancement
The arcs and circles of the slight structures seem to sharpen when closing the eyes a little bit. By squeezing the eyes, one reduces the amount of the incident light rays. One also has the impression that noise is added to the picture we see (in this context the slight twitches of the eyelid muscles and/or the light diffraction through the eyelashes may play a role, too). The brain interprets the incoming information and suggests us a visual perception of a sharpened image. The event of adding noise to a weak signal to enhance it in some way is similar to a physical phenomenon called stochastic resonance.
Stochastic resonance
Stochastic resonance (SR) denotes the occurrence, that weak signals of a non-linear system can be enhanced detected when adding noise to the system. This effect is used e.g. in signal analysis or in medical devices to enhance sensory and motor function. Often the signal is threshold filtered after adding noise. SR also occurs naturally in the sensory systems of several organisms.
Here we want to show the idea of stochastic resonance with an image detail out of chromosome 17 (see Fig. 1). Therefore we need a signal, additive noise and a threshold.
The “signal” is the image detail converted to a 0 to 255 gray scale (Fig. 2). As “noise” there is added a random number j, created from a Gaussian distribution with zero mean and standard deviation σ, to each gray value i in every pixel of the picture. The result is compared to a threshold value T. If the new gray value is below the threshold value ( j + i < T), the gray value in that pixel is replaced with 0 (black). If the new gray value crosses the threshold ( j + i > T), the pixel is marked with 255 (white).

Fig. 1: Detail of chromosome 17

Fig. 2: Conversion into gray scale
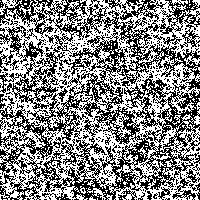
Fig. 3: Single SR-image
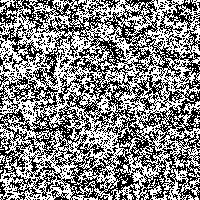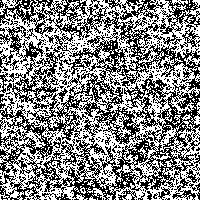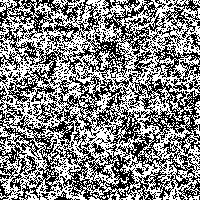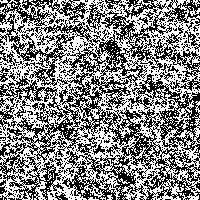
Fig. 4: Series of SR-images
The possibility to adjust the most suitable noise intensity for the image is given by the two parameters standard deviation σ and threshold T. For low noise intensities very few signal crosses the threshold, leading to a low signal-to-noise ratio without perceptive enhancement. In contrast, large noise intensities cause a swamp of the threshold by the noisy signal, as well leading to a low signal-to-noise ratio without perceptive enhancement. But there exists an optimum of noise intensity which allows the signal to cross the threshold moderately (high signal-to-noise ratio) and therefore leading to a perceived image enhancement.
To get to know the influence of the different parameters on the image rendered with stochastic resonance we created a tool with which you can play around with the values of the standard deviation σ, threshold T, and the time gaps between the images of the animation strip (delay).
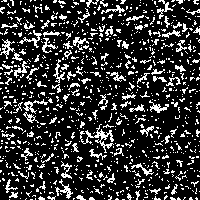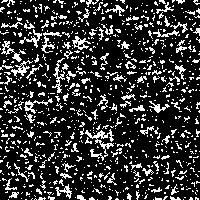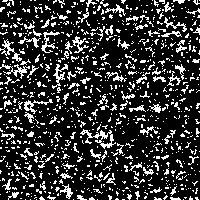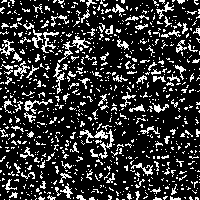
With this example we just want to show the effect of stochastic resonance applied to an image detail of our rendered DNA-images. If it makes any scientific sense to extract those slight structures remains unclear.
